by Aaron Jonas Stutz
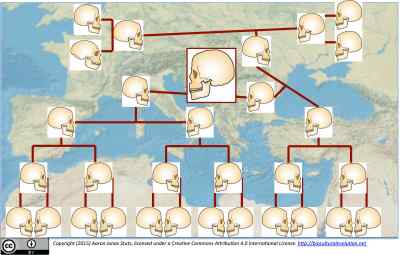
The Nature publishing group has taken a welcome step toward making the details of scientific research more widely accessible. The flagship cross-disciplinary journal Nature now provides full PDF text views for published articles linked from major media sites. Thus, the article on ancient genetically admixed Neandertal and modern human DNA recovered from the Oase I mandible–work authored by Qiaomei Fu and colleagues–has now been published, and everyone can read the full-text on nature.com. It’s available to everyone via media outlets such as the BBC here. You can click through the hyperlinked text in the BBC story. Now, you can’t download the PDF, but you can view it (apparently only once, as I discovered in trying to verify the BBC link just now) in its entirety in your web browser. It’s not open access, but it’s “opener.”
So what about the full, published version?
It’s been much anticipated, since Fu reported on her work at a recent genomics conference in Cold Springs Harbor (Callaway, 2015; Gibbons, 2015).
And, well, there are no major surprises to add to the main take-home from the Cold Springs Harbor conference presentation. The quite anatomically modern mandible, labelled Oase I, from the Peştera cu Oase in Romania, dates to ca. 42-39 thousand years ago, and the extraction and analysis of DNA fragments preserved in the mandible’s subfossil bone tissues indicate that this male individual had a very recent, very Neandertal ancestor, four to six generations back. I wrote in detail about the implications of this finding in the post Why None of Us Are Very Neandertal–But Most of Us Are a Little.
Still, in the full article (Fu et al., 2015), there are some additional points worth highlighting:
1) The biochemical techniques for capturing huge libraries of fragmented bits of DNA just continue improve. For us non-genomics-specialists who work on related methods and sources of information—that is, for archaeologists like me—this is huge. OK. The details are that Fu et al. (2015) successfully aligned really small extracted DNA fragments with known sequences in each human nuclear chromosome and the mitochondrial DNA. And this only amounted to a fraction of one percent of the entire three-billion-base-pair genome. Still, it included around two million fragments long enough to be aligned with loci encompassing single-nucleotide positions that are known to vary in living human populations. Screening for chemical markers of post mortem contamination or alteration reduced this number to around 200,000 single nucleotide polymorphism sites. Further cautious screening identified close to 80,000 single nucleotide polymorphism loci (SNPs) whose locations are known on the autosomal chromosomes (our 22 pairs of non-sex chromosomes) in the human genome map, exhibiting different alleles among ancient and modern human genomes. That gives us a respectable, although quite low-resolution scan of the entire genome. Additional SNPs from the sex chromosomes are what confirm that Oase I has both an X and a Y chromosome. All of this recovery of very fragmentary genomic data is important, because Oase I simply isn’t as well-preserved as many other roughly contemporaneous hominin specimens–including those representing archaic Asians (a.k.a. Denisovans), archaic western Eurasians (a.k.a. Neandertals), and early anatomically modern human Eurasians. Along with the recent publication of work aiming to push the limits of extracting uncontaminated bits of DNA from poorly chemically and histologically preserved bone (Pinhasi et al., 2015), the new report by Fu and colleagues (2015) will encourage serious work on extracting DNA from fossils in warmer, wetter environments–where we thought ancient bones simply couldn’t preserve intact genomic material from the original biological tissues.
2) Oase I probably didn’t just have one very recent invidivual Neandertal ancestor, which was likely a great-great or great-great-great grandparent. This very recent ancestry is inferred by observing the linked proximity of many of the SNP loci that have Neandertal alleles. The DNA fragmentation notwithstanding, Fu et al. (2015) recovered enough non-damaged DNA to align with the complete human genome map, and a surprisingly large number of the Oase SNPs with Neandertal alleles are from loci close to one another on virtually every chromosome. The authors write that “the Neanderthal contribution to the Oase 1 individual occurred so recently in his family tree that chromosomal segments of Neanderthal origin had little time to break up due to recombination.”
However, they continue:
When we remove the seven longest segments, the estimate of Neanderthal ancestry in Oase 1 drops from 7.3% to 4.8%, which is still around twice the 2.0–2.9% estimated for the French, Han, Kostenki and Ust’-Ishim individuals in this remaining part of the genome. This additional Neanderthal ancestry could reflect an older Neanderthal admixture into the ancestors of Oase 1, or that we failed to find all segments of recent Neanderthal ancestry.
The substantially older Ust-‘Ishim anatomically modern human (from ca. 45,000 years ago) inherited numerous Neandertal alleles from a range of earlier ancestors who had admixed with African populations expanding into western Eurasia, perhaps 60-50 thousand years ago (Fu et al., 2014). So, it’s likely that Oase I also had some of these older Neandertal ancestors, too. The Oase I data support investigating the hypothesis that there were multiple episodes of admixture, and that later ones had only geographically patchy effects.

3) But still, we have to remember that the Oase I aligned, non-damaged DNA fragments are mainly non-Neandertal. Let’s compare Oase I with the handfull of other, roughly contemporary or slightly later Eurasian anatomically modern human bones from whom researchers have recovered largely intact genomic information. Among these individuals, Oase I is clearly the most closely related to other roughly contemporary or slightly older Neandertals for whom genomic data is also available. In other words, Oase I has the most recent Neandertal ancestors. But at the same time, my point is that Oase I still has mainly anatomically modern, recent African ancestry … just like the rest of us living today. This is evidence that the scale of biological turnover–in which Neandertal anatomical patterns were nearly fully replaced by anatomically modern ones–was thoroughly extensive in Eurasia. In fact, Fu and colleagues (2015) confirm what the same team had recently shown in analyzing better-preserved, later western Eurasian anatomically modern humans–dating from roughly 35,000 to 5,000 years ago. The process of turnover in western Eurasia, in particular, progressively continued through the timeframe ca. 40-35,000 years ago, as anatomically modern human metapopulations–that is, sets of neighboring regional populations interconnected by regular but restricted migration and interbreeding–became well-established across Eurasia and evolved a degree of isolation by distance, with distinctive alleles at polymorphic sites subtly but reliably differentiating western and eastern Eurasian Upper Paleolithic genomes (Lazaridis et al., 2014). Interestingly, the whole process of complex, recurrent admixture seems to have happened all over again with population expansion at the beginning of the modern warm period, the Holocene, especially in connection with agricultural expansion in western Eurasia (Lazaridis et al., 2014). This complex history of population range expansion and admixture seems to have occurred repeatedly in human evolution. As I have emphasized in my earlier post, Alan Templeton drew this conclusion from more limited genetic data already many years ago (Templeton, 2002). But we can now trace the details much more thoroughly and accurately, thanks to the recovery of ancient genomic-scale information from very old human bones.
Anatomically Modern Human-Neandertal Turnover: Selection within Homo sapiens or Competition between Species?
One hope I have for near-future genomics work is this. There should now be enough genomic detail, perhaps even from Oase I, to look at whether there are different patterns of linkage disequilibrium in Neandertals and anatomically modern humans from this time frame, ca. 50,000-35,000 years ago. It is within this period that most of the really well-preserved Neandertal genome sequences, along with the Ust-‘Ishim, Kostenki XIV, and Oase I western Eurasian anatomically modern humans, are all known to date (Fu et al., 2015).
Remember that linkage disequilibrium occurs when you get long stretches of DNA inherited from a given parent or grandparental ancestor intact, not randomly mixed with alleles from other, older recent ancestors during crossing over in meiosis. And in the current article (Fu et al., 2015), linkage disequilibrium patterns constitute strong evidence for a very recent Neandertal having a place in a particular anatomically modern guy’s family tree.
But as Hawks et al. (2007) have shown, linkage disequilibrium patterns in living human genomes can provide evidence for something else!!! Rapid natural selection can keep unusually long lengths of DNA together in people across an entire metapopulation. At least after a number of generations.
This is how it works. Those individuals with a fitness-enhancing functional allele–whether it has to do with coding for proteins or regulating RNA function in the immune system, digestion, growth, brain activity, aging, or whatever–will be marginally more likely to survive and send more offspring with that allele into the next generation. In each literal instance of reproductive success, the fitness-enhancing DNA locus in that environment would carry with it linked bits of non-functional DNA, which we definitely have lots of. The more rapid the natural selection–and as Hawks et al., (2007) emphasize, a rapid intergenerational increase in the population frequency of a given allele may not have to do with strong fitness differentials among extant alleles, but rather simply a larger biocultural context of population expansion, which carries with it larger absolute numbers of mutations and greater intrinsic population variation in reproductive success, heightening the potential impact of natural selection–the more individuals inherit long chunks of linked non-coding bits around the favored allele. And thus, the stronger the population-level trace of linkage disequilibrium, generations later.
Now, if natural selection extensively shaped the process of anatomically modern human-Neandertal turnover, we should see sets of linked SNPs in early anatomically modern human Eurasians that are loci within or next to known genes or important regulatory regions… and we shouldn’t see these linked SNP markers in Neandertals. Researchers have been interested in Neandertal alleles introgressing into anatomically modern human populations–and then becoming favored by natural selection (Abi-Rached et al., 2011; Hawks & Cochran, 2006; Racimo et al., 2015). Already, Sankararaman and colleagues (2014) have compared well-preserved ancient Neandertal genomes to those of over 1000 living humans, and they discovered that in the modern genomes, several regions rich in protein-coding loci in the autosomal chromosomes and the X chromosome were nearly devoid of Neandertal ancestry … whereas the rest of the Asian and European sample genomes had a background of 1-3% Neandertal ancestry. This is very strong evidence that in admixed Neandertal-anatomically modern human populations, there was a whole range of functional Neandertal alleles that caused some degree of “hybrid disadvantage.” Sankararaman et al. (2015) present a plausible argument for investigating the hypothesis that X-linked male sterility–or at least, lower fertility–played a role in selection against Neandertal genes. I emphasize that this is plausible, precisely because Neandertal metapopulations were substantially smaller over their evolutionary history than were the anatomically modern African ones that sent out the demographic growth-and-dispersal waves into Eurasia. With low initial diversity at around 400-200 thousand years ago and possible initial period of geographic isolation between Neandertals and other populations, the nature of genetic inheritance is such that mutations affecting changes in gamete compatibility could really snowball over a period of generations (see Städler et al., 2012 for an important recent study of how hybrid sterility evolves in plants). Thus, we really need to see if–in the recently reconstructed genomes from Early Upper Paleolithic anatomically modern humans–there’s a linkage disequilibrium signal of this kind of selection going on during the period of biological turnover.
We expect many anatomically modern human alleles to have been favored in the face of recurrent admixture with Neandertals in Eurasia. Although male sperm function or gamete compatibility may be one factor, some other predicted loci under selection should be haplotypes that influenced fitness favoring observable or inferred anatomically modern phenotypes. As I discussed in my previous post on Oase I, such phenotypes have been variously proposed to include life-history (maturation and aging-rate) traits, body-size and body-proportion traits, craniofacial traits related to chewing and mechanical dental loading, craniofacial traits related to oral and facial expression visibility, and frontal morphology and its possible relationship to neocortical neural organization.
And the reason that we should be interested in tracing the role of natural selection in anatomically modern human-Neandertal turnover is that, while Early Upper Paleolithic populations in Eurasia experienced a slight bump in population size, this was so small that anatomically modern humans definitely did not demographically swamp Neandertal populations. There was likely something about the Early Upper Paleolithic Eurasian biocultural environment—perhaps involving a marginal decline in residential move distances and daily foraging rounds, along with a marginal increase in length of stay at some residential camps and the size of co-residential and task groups—that favored anatomically modern phenotypes that had been evolving in Africa for tens of thousands of years already.
This brings me back to the question of whether anatomically modern humans and Neandertals constituted distinct biological species. The evidence from Oase I suggests on its face, no. There are many stable population systems in which closely related metapopulations are separated by what seems to be a resilient hybrid zone. Yet, Oase I is clearly not an instance of such hybridization. Oase I’s ancestry was shaped by a context in which anatomically modern humans underwent population-geographic range expansion out of Africa, and this conspicuously involved recurrent admixture with Neandertal groups in Eurasia.
But … as Sankararaman and colleagues (2014) argue, this doesn’t mean a speciation model doesn’t fit here.
Indeed, earlier episodes of random genetic drift, geographic isolation, and millennia of adaptation to distinct biocultural niches arguably shaped an incipient process of speciation—one that hadn’t resulted in a well-defined reproductive barrier.
As niche construction and adaptation resulted in the emergence of a (slightly) lower-mobility, higher-density, higher-social-connectivity human niche in Africa, anatomically modern human populations would have sent recurrent (albeit quite low-level) waves of geographic dispersal into Eurasia. Such a multiple-dispersal scenario has recently been put forward by Boivin and colleagues (2013). It is possible that the more socially intense niche—which is well-attested to in parts of Africa, at various times and places, during the Middle Stone Age period (McBrearty & Brooks, 2000)—shaped independent adaptations among admixed Neandertal-anatomically modern human groups just outside of Africa. All that is necessary for Eurasian Upper Paleolithic biocultural adaptations is a niche with higher population density and greater social network connectivity. Whether this was also sufficient … well, that’s not so clear. It may alternatively be that biocultural adaptations constituting Early Upper Paleolithic populations, technologies, and societies were modifications resulting from socially-structured dispersal from Africa. In other words, African groups may have carried with them a cultural inheritance that was then modified in the Levant or south Asia and then modified in various ways across Eurasia after 50,000 years ago. And all of this may have led to a selective process of reinforcement—but in an unusual context of one emerging species having evolved an altered social niche that favored anatomically modern traits over Neandertal ones.

I have to be honest here. I can’t see this particular debate getting resolved any time soon. We’re talking about whether there’s a speciation process marked by more rapid social niche construction emerging in the geographic center of the genus’s range … or whether the metapopulations within a single species’s geographic center are involved in niche construction that then shapes expansion and replacement with admixture and selection at the edges. The two scenarios aren’t so different. It’s a matter of where you draw the boundary between population divergence and reinforcement versus metapopulation dynamics with lots of natural selection.
I’m for the latter, but only on the balance. It’s interesting to think about how recent evolution in the genus Homo may have followed many of the same population geographic dynamics involved in allopatric speciation in non-human cases. But an important part of human uniqueness lies in the recurrent emergence of more complex, organized, extensive social niches. And that makes Neandertal extinction difficult to compare to clearer examples of reinforcement and competitive exclusion between closely related species in the same genus.
References
Abi-Rached, L., Jobin, M. J., Kulkarni, S., McWhinnie, A., Dalva, K., Gragert, L., … Parham, P. (2011). The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans. Science, 334(6052), 89–94. http://doi.org/10.1126/science.1209202
Boivin, N., Fuller, D. Q., Dennell, R., Allaby, R., & Petraglia, M. D. (2013). Human dispersal across diverse environments of Asia during the Upper Pleistocene. Quaternary International, 300, 32–47. http://doi.org/10.1016/j.quaint.2013.01.008
Callaway, E. (2015). Early European may have had Neanderthal great-great-grandparent. Nature. http://doi.org/10.1038/nature.2015.17534
Fu, Q., Hajdinjak, M., Moldovan, O. T., Constantin, S., Mallick, S., Skoglund, P., … Pääbo, S. (2015). An early modern human from Romania with a recent Neanderthal ancestor. Nature, advance online publication. http://doi.org/10.1038/nature14558
Fu, Q., Li, H., Moorjani, P., Jay, F., Slepchenko, S. M., Bondarev, A. A., … Pääbo, S. (2014). Genome sequence of a 45,000-year-old modern human from western Siberia. Nature, 514(7523), 445–449. http://doi.org/10.1038/nature13810
Gibbons, A. (2015). Ancient DNA pinpoints Paleolithic liaison in Europe. Science, 348(6237), 847–847. http://doi.org/10.1126/science.348.6237.847
Hawks, J., & Cochran, G. (2006). Dynamics of Adaptive Introgression from Archaic to Modern Humans. PaleoAnthropology, 2006, 101–115. http://www.paleoanthro.org/media/journal/content/PA20060101.pdf
Hawks, J., Wang, E. T., Cochran, G. M., Harpending, H. C., & Moyzis, R. K. (2007). Recent acceleration of human adaptive evolution. Proceedings of the National Academy of Sciences, 104(52), 20753–20758. http://doi.org/10.1073/pnas.0707650104
Hvala, J. A., & Wood, T. E. (2001). Speciation: Introduction. In eLS. John Wiley & Sons, Ltd. Retrieved from http://onlinelibrary.wiley.com.proxy.library.emory.edu/doi/10.1002/9780470015902.a0001709.pub3/abstract
Lazaridis, I., Patterson, N., Mittnik, A., Renaud, G., Mallick, S., Kirsanow, K., … Krause, J. (2014). Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature, 513(7518), 409–413. http://doi.org/10.1038/nature13673
Mcbrearty, S., & Brooks, A. S. (2000). The revolution that wasn’t: a new interpretation of the origin of modern human behavior. Journal of Human Evolution, 39(5), 453–563. http://doi.org/10.1006/jhev.2000.0435
Pinhasi, R., Fernandes, D., Sirak, K., Novak, M., Connell, S., Alpaslan-Roodenberg, S., … Hofreiter, M. (2015). Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PLoS ONE, 10(6), e0129102. http://doi.org/10.1371/journal.pone.0129102
Racimo, F., Sankararaman, S., Nielsen, R., & Huerta-Sánchez, E. (2015). Evidence for archaic adaptive introgression in humans. Nature Reviews Genetics, 16(6), 359–371. http://doi.org/10.1038/nrg3936
Sankararaman, S., Mallick, S., Dannemann, M., Prüfer, K., Kelso, J., Pääbo, S., … Reich, D. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature, 507(7492), 354–357. http://doi.org/10.1038/nature12961
Städler, T., Florez-Rueda, A. M., & Paris, M. (2012). Testing for “Snowballing” Hybrid Incompatibilities in Solanum: Impact of Ancestral Polymorphism and Divergence Estimates. Molecular Biology and Evolution, 29(1), 31–34. http://doi.org/10.1093/molbev/msr218
Templeton, A. (2002). Out of Africa again and again. Nature, 416(6876), 45–51. http://doi.org/10.1038/416045a